Chemical Cross-Linkers
Most proteins act in association with other proteins to form protein complexes stably or transiently in cells, and mapping these interactions is essential to understand their cellular functions. Protein complexes represent functional entities that are often difficult to analyze using conventional structural tools due to their heterogeneous and dynamic nature. Recently, cross-linking Mass Spectrometry (XL-MS) has been recognized as a valuable tool for the structural analysis of protein assemblies, which can be used alone and in combination with other techniques. In addition to in vitro studies, XL-MS approaches have been extended to capture protein interactions in living cells. Identification of cross-linked peptides by MS analysis can provide distance constraints to assist computational modeling and yield structural information at amino acid resolution. The advantages of cross-linking studies include small sample size, robust tolerance for size and environment of the protein complex, instrument accessibility, and the speed of handling and data collection.
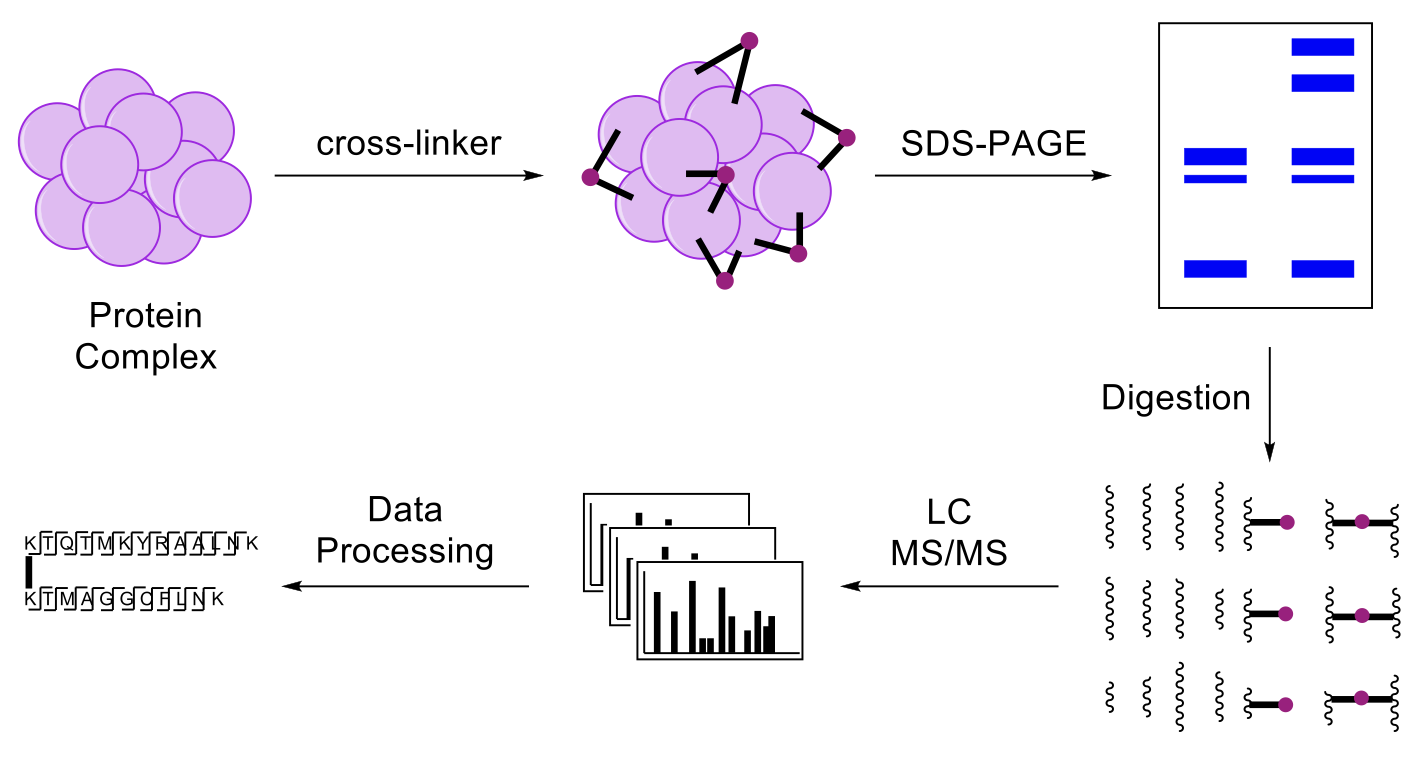
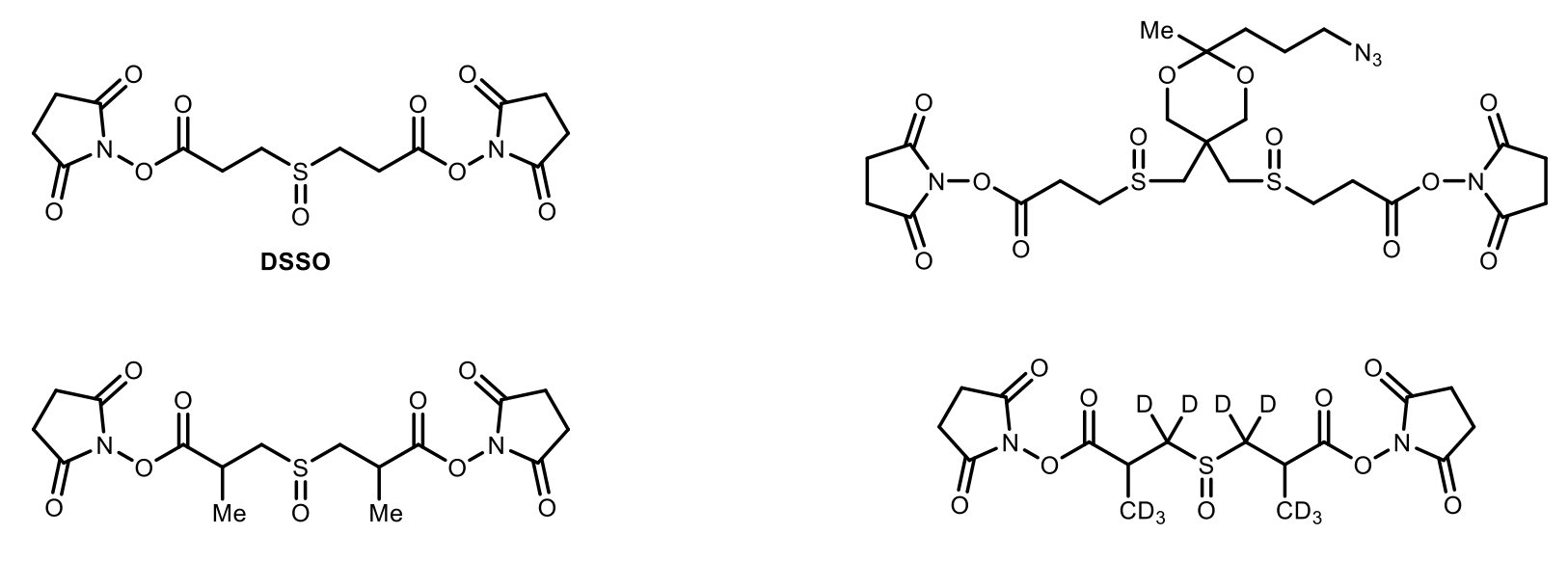
Unambiguous identification of cross-linked peptides can be greatly facilitated by the introduction of a MS cleavable bond in a cross-linking reagent, which can fragment during collision induced dissociation (CID) prior to peptide backbone breakage. Previously, we have successfully developed a new class of robust MS-cleavable reagents that contain labile C–S sulfoxide bonds (e.g. DSSO (disuccinimidylsulfoxide)), and thus enables fast and accurate identification of cross-linked peptides using liquid chromatography-multistage tandem mass spectrometry analysis (LC/MS).