From: Treseder KK, Lennon JT. 2015. Fungal traits that drive ecosystem dynamics on land. Microbiology and Molecular Biology Reviews 79:243-262.
Under cold conditions, RNAs can form stable tertiary structures that render them non-functional and prevent translation (Owttrim 2006). Certain cold-induced RNA helicases can unwind the RNAs or bind to them, which allows translation to proceed (Jones et al. 1996, Chamot et al. 1999). Fungi that carry these RNA helicases display improved cold tolerance (Strauss and Guthrie 1991, Noble and Guthrie 1996, Lim et al. 2000, Schade et al. 2004, Ellison et al. 2011, Markkula et al. 2012), and can be more prevalent in colder environments (Ellison et al. 2011). RNA helicase may form part of a generalized stress response (Gasch and Werner-Washburne 2002, Schade et al. 2004).
Fungal genes for cold-induced RNA helicase
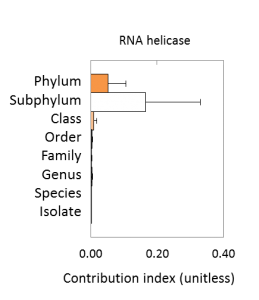
Genetic capacity for cold-induced RNA helicases varies most at the subphylum level:
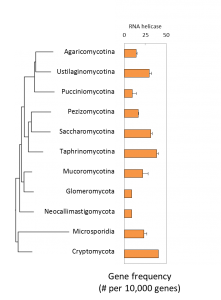
Phylogenetic distribution of genes for cold-induced RNA helicases:
Yeasts have the highest abundance of genes for cold-induced RNA helicases:
Chamot, D., W. C. Magee, E. Yu, and G. W. Owttrim. 1999. A cold shock-induced cyanobacterial RNA helicase. Journal of Bacteriology 181:1728-1732.
Ellison, C. E., C. Hall, D. Kowbel, J. Welch, R. B. Brem, N. L. Glass, and J. W. Taylor. 2011. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci U S A 108:2831-2836.
Gasch, A. P. and M. Werner-Washburne. 2002. The genomics of yeast responses to environmental stress and starvation. Funct Integr Genomics 2:181-192.
Jones, P. G., M. Mitta, Y. Kim, W. N. Jiang, and M. Inouye. 1996. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci U S A 93:76-80.
Lim, J., T. Thomas, and R. Cavicchioli. 2000. Low temperature regulated DEAD-box RNA helicase from the Antarctic archaeon, Methanococcoides burtonii. Journal of Molecular Biology 297:553-567.
Markkula, A., M. Lindstrom, P. Johansson, J. Bjorkroth, and H. Korkeala. 2012. Roles of four putative DEAD-Box RNA helicase genes in growth of Listeria monocytogenes EGD-e under heat, pH, osmotic, ethanol, and oxidative stress conditions. Appl Environ Microbiol 78:6875-6882.
Noble, S. M. and C. Guthrie. 1996. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics 143:67-80.
Owttrim, G. W. 2006. RNA helicases and abiotic stress. Nucleic Acids Research 34:3220-3230.
Schade, B., G. Jansen, M. Whiteway, K. D. Entian, and D. Y. Thomas. 2004. Cold adaptation in budding yeast. Molecular Biology of the Cell 15:5492-5502.
Strauss, E. J. and C. Guthrie. 1991. A cold-sensitive messenger RNA splicing mutant is a member of the RNA helicase gene family. Genes & Development 5:629-641.