Research
RNA at the Center of Cancer Signaling & Metabolism
RNA is a crucial genetic material that acts at the central node in gene expression programs. The Lee lab is dedicated to uncovering the involvement of post-transcriptional quality control of RNA molecules, including splicing, chemical modification, transport, stabilization and translation, in rewiring of cellular signaling and metabolism during tumorigenesis. Our ultimate goal is to develop therapeutic approaches and diagnostic tools targeting RNA and metabolic vulnerabilities of cancers.
Kidney Cancer
The kidney is a vital organ responsible for eliminating waste products from the blood while reabsorbing essential nutrients back into the body. Our research focuses on malignant tumor formation in the kidneys, as these tumors not only disrupt kidney function but also have far-reaching consequences on the metabolism of the entire body. Our particular area of interest lies in studying a genetic kidney tumor syndrome known as Tuberous Sclerosis Complex (TSC). This syndrome arises from loss-of-function mutations in the TSC1/2 tumor suppressor genes, leading to hyperactivation of the mechanistic target of rapamycin (mTORC1) signaling, a master regulator of cell growth and proliferation that is overactivated in most human cancers.
Using metabolomic, transcriptomic, and proteomic analyses of genetic and xenograft mouse models, patient-derived cell lines, and patient tissues and serum samples, we elucidate the molecular mechanisms involved in kidney tumor pathogenesis. We found that TSC tumors exhibit overactive lipid metabolism due to the increased expression of fat synthesizing enzymes. Mechanistically, mTORC1 induces the expression of these metabolic enzymes by promoting their RNA splicing and stability. On the other hand, inhibition of these fat synthesis enzymes, combined with extracellular lipid depletion, attenuates TSC tumor growth (Cell 2017; Molecular Cell 2023). Our findings have brought to light the critical role of mTORC1 signaling and RNA processing in the metabolic adaptation of cancer cells, enhancing RNA splicing, stability, and translation of vital metabolic enzymes. Through our research efforts in kidney and other types of human cancers, we hope to contribute to the development of effective treatments and diagnostic tools that can enhance outcomes for cancer patients.
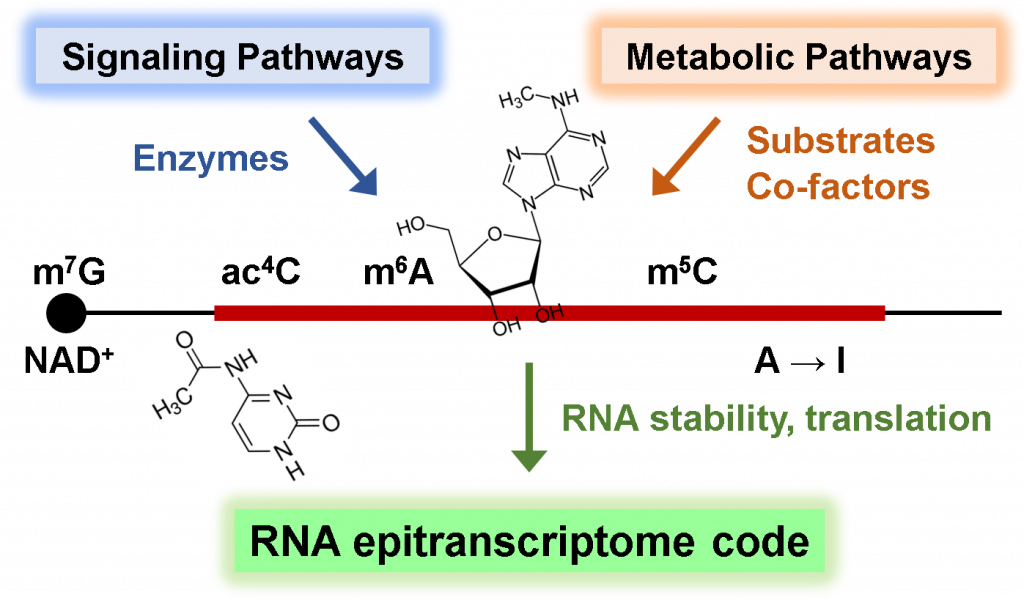
Epigenetic Modification of RNA
We also explore the fascinating world of RNA epigenetics, where chemical modifications occur on the sugars and bases of RNA molecules. Like DNA and histones undergoing epigenetic modifications, RNA contains an astounding repertoire of over 150 diverse modifications, suggesting the existence of intricate regulatory mechanisms and their crucial functions in gene expression regulation and cell fate decisions. By integrating liquid chromatography-mass spectrometry (LC-MS), RNA modification sequencing, and mathematical modeling, along with in-depth molecular biological and biochemical mechanistic studies, we investigate the signaling and metabolic pathways that control RNA chemical modification, as well as the crosstalk between RNA modification and cellular metabolic processes in normal and malignant cells.
Our research has already yielded exciting results, including our recent discovery of growth factor and oncogenic signaling induced N6-methyl-adenosine (m6A) mRNA modification, and drug resistance mechanisms by m6A-dependent gene expression program (Molecular Cell 2021; JBC 2023). Through our research, we aim to delineate the hidden signaling and metabolic codes embedded within RNA molecules, define how these epigenetic chemical modifications contribute to gene expression programs, and elucidate how their dysregulation leads to human diseases.
Selected Publications
(Lee lab trainees; *Equal contribution; #Corresponding author)
2024
- Chun Y, Fruman DA, Lee G#. The picky mTORC1 in metabolic enzyme degradation. Molecular Cell (2024). [PDF]
- Yang M, Zheng X, Fan J, Cheng W, Yan T, Lai Y, Zhang N, Lu Y, Qi J, Huo X, Xu Z, Huang J, Jiao Y, Liu B, Pang R, Zhong X, Huang S, Luo G, Lee G, Jobin C, Eren AM, Chang EB, Wei H, Pan T, Wang X. Antibiotic-induced gut microbiota dysbiosis modulates host transcriptome and m6A epitranscriptome via bile acid metabolism. Advanced Science (2024) 2307981.
- Rubtsova VI, Chun Y, Kim J, Ramirez CB, Jung S, Cassidy E, Rushing G, Aguiar DJ, Lau WL, Ahdoot RS, Smith M, Lee S, Jang C, Lee G#. Circulating biomarkers of kidney tumors in TSC (tuberous sclerosis complex) patients. medRxiv (2024). [PDF]
2023
- Nguyen TB, Miramontes R, Chillon-Marinas C, Maimon R, Vazquez-Sanchez S, Lau AL, McClure NR, England WE, Singha M, Stocksdale JT, Jang K, Jung S, McKnight JI, Ho LN, Faull RLM, Steffan JS, Reidling JC, Jang C, Lee G, Cleveland DW, Lagier-Tourenne C, Spitale RC, Thompson LM. Aberrant splicing in Huntington’s disease via disrupted TDP-43 activity accompanied by altered m6A RNA modification. bioRxiv (2023).
- Kim J, Chun Y, Ramirez CB, Hoffner LA, Jung S, Jang K, Rubtsova VI, Jang C, Lee G#. MAPK13 stabilization via m6A modification limits anti-cancer efficacy of rapamycin. Journal of Biological Chemistry (2023) 299(9):105175. [PDF]
- Preprint: Kim J, Chun Y, Ramirez CB, Hoffner LA, Jung S, Jang K, Rubtsova VI, Jang C, Lee G#. MAPK13 stabilization via m6A modification limits anti-cancer efficacy of rapamycin. bioRxiv (2022).
- Cho S, Chun Y, He L, Ramirez CB, Ganesh KS, Jeong K, Song J, Cheong JG, Li Z, Choi J, Kim J, Koundouros N, Ding F, Dephoure N, Jang C, Blenis J#, Lee G#. FAM120A couples SREBP-dependent transcription and splicing of lipogenesis enzymes downstream of mTORC1. Molecular Cell (2023). [PDF]
2022
- Yaron TM, Heaton BE, Levy TM, Johnson JL, Jordan TX, Cohen BM, Kerelsky A, Lin T, Liberatore KM, Bulaon DK, Nest SJV, Koundouros N, Kastenhuber ER, Mercadante MN, Shobana-Ganesh K, He L, Schwartz RE, Chen S, Weinstein H, Elemento O, Piskounova E, Nilsson-Payant BE, Lee G, Trimarco JD, Burke KN, Hamele CE, Chaparian RR, Harding AT, Tata A, Zhu X, Tata PR, Smith CM, Possemato AP, Tkachev SL, Hornbeck PV, Beausoleil SA, Anand SK, Aguet F, Getz G, Davidson AD, Heesom K, Kavanagh-Williamson M, Matthews D, tenOever BR, Cantley LC, Blenis J, Heaton NS. Host protein kinases required for SARS-CoV-2 nucleocapsid phosphorylation and viral replication. Science Signaling (2022).
- Jang K, Heras CR, Lee G#. m6A in the signal transduction network. Mol Cells (2022). [PDF]
2021
- Mathur L*, Jung S*, Jang C#, Lee G#. Quantitative analysis of m6A RNA modification by LC-MS. STAR Protocols (2021). [PDF]
- Cho S*, Lee G*#, Pickering BF*, Jang C, Park J, He L, Mathur L, Kim S, Jung S, Tang H, Monette S, Rabinowitz JD, Perrimon N, Jaffrey SR#, Blenis J#. mTORC1 promotes cell growth via m6A-dependent mRNA degradation. Molecular Cell (2021). [PDF]
- Commentary by Michael N. Hall (Lasker Award winner): “More writing: mTORC1 promotes m6A mRNA methylation” Molecular Cell (2021).
- Tang HW, Weng JH, Lee WX, Hu Y, Gu L, Cho S, Lee G, Binari R, Li C, Cheng ME, Kim AR, Xu J, Shen Z, Xu C, Asara JM, Blenis J, Perrimon N. mTORC1-chaperonin CCT signaling regulates m6A RNA methylation to suppress autophagy. PNAS (2021).
- Kim J, Lee G#. Metabolic control of m6A RNA modification. Metabolites (2021). [PDF]
Before 2020
- Park JH, Lee G, Blenis J. Structural insights into the activation of mTORC1 on the lysosomal surface. Trends Biochem Sci. (2020).
- Krishnamoorthy GP, Davidson NR, Leach SD, Zhao Z, Lowe SW, Lee G, Landa I, Nagarajah J, Saqcena M, Singh K, Wendel HG, et al. EIF1AX and RAS mutations cooperate to drive thyroid tumorigenesis through ATF4 and c-MYC. Cancer Discovery (2019).
- Zheng Y, Lin TY, Lee G, Paddock MN, Momb J, Cheng Z, Li Q, Fei DL, Stein BD, Ramsamooj S, Zhang G, Blenis J, Cantley LC. Mitochondrial one-carbon pathway supports cytosolic folate integrity in cancer cells. Cell (2018). [PDF]
- He L, Gomes AP*, Wang X*, Yoon SO*, Lee G, Nagiec M, Cho S, Chavez A, Islam T, Yu Y, Asara JM, Couvillon A, Kim BY, Blenis J. mTORC1 promotes metabolic reprogramming by suppression of Foxk1 phosphorylation. Molecular Cell (2018).
- Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, Yu Y, Liu X, He L, Cavaliere PM, Chavez A, Zhang E, Isik M, Couvillon A, Dephoure NE, Blackwell TK, Yu JJ, Rabinowitz JD, Cantley LC, Blenis J. Post-transcriptional regulation of de novo lipogenesis by mTORC1-S6K1-SRPK2 signaling. Cell (2017). [PDF]
- Commentary: “Intron splicing for lipid biosynthesis” Science Signaling (2018).
- Commentary: “SRPK2 acts downstream of mTORC1 to promote de novo lipogenesis” Cancer Discovery (2018).
- Csibi A*, Lee G*, Yoon SO, Tong H, Ilter D, Elia I, Fendt SM, Roberts TM, Blenis J. The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B dependent control of c-Myc translation. Current Biology (2014).
- Lee G, Blenis J. Akt-ivation of RNA splicing. Molecular Cell (2014).
- Kim H, Lee JM*, Lee G*, Bhin J*, Oh SK, Kim K, Pyo KE, Lee JS, Yim HY, Kim KI, Hwang D, Chung J, Baek SH. DNA damage-induced RORα is crucial for p53 stabilization and increased apoptosis. Molecular Cell (2011).
- Sun D*, Lee G*, Lee JH*, Kim H, Rhee H, Park S, Kim K, Kim Y, Kim BY, Hong J, Park C, Choy HE, Kim JH, Jeon YH, Chung J. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nature Structural & Molecular Biology (2010).
- Hyun S*, Lee JH*, Jin H*, Nam JW, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell (2009).
- Lee G, Chung J. Discrete functions of rictor and raptor in cell growth regulation in Drosophila. Biochem. Biophys. Res. Commun. (2007).
Complete List
Members
Gina Lee, PhD (Principal Investigator) (Faculty Profile)
Assistant Professor at UC Irvine School of Medicine. Gina got postdoctoral training at Harvard Medical School / Weill Cornell Medicine of Cornell University, and BS and PhD from KAIST / Seoul National University. She is a recipient of Breakout prize, NIH-NCI career transition award, and Mary Kay Ash cancer research grant. She likes walking, jogging, reading books, coffee, and chocolate.
Izabelle Le, BS (Lab Manager/Research Assistant)
Izabelle graduated from UCSD with a BS in Biochemistry. She is interested in and enjoys recombinant DNA and RNA technology and learning about cancer metabolism. When she’s not in the lab, Izabelle spends her time dancing, crocheting, watching movies, and coffee shop hopping. Although STEM and research are her main pursuits, she often dabbles in the artistic side of herself.
Joohwan Kim, PhD (Postdoctoral Fellow)
During his PhD at Kangwon National University School of Medicine, Joohwan studied tumor angiogenesis and microRNA biogenesis. He received TSC Alliance postdoctoral fellowship and Vicky Whittemore travel award for his RNA epigenetics research in TSC kidney tumors. While Joohwan’s passion lies in scientific endeavors, he also loves Donkatsu and enjoys meditation.
Ki-Hong Jang, PhD (Postdoctoral Fellow)
Ki-Hong focused on studying the comprehensive cell death signaling cascades during his PhD at Chungnam National University. Currently, he delves into the realm of cell survival signaling and metabolism through RNA chemical modification. Aside from his dedication to science, Ki-Hong’s love for basketball becomes evident as he skillfully takes on the roles of shooting guard and small forward.
Yujin Chun, PhD (Postdoctoral Fellow)
Yujin completed her PhD at KAIST, where she dedicated her research to unraveling nuclear exosomes and 3D chromatin structures. She has expertise in molecular genetics and bioinformatics and aims to expand her research strength into human cancer and metabolic diseases during her postdoctoral training. Yujin enjoys indoor climbing and spending free time with her dogs. She received a postdoctoral fellowship from the National Research Foundation of Korea.
Cuauhtemoc (Temoc) Ramirez, BS (PhD Student)
Temoc is a PhD student co-mentored by Dr. Cholsoon Jang. He is interested in the biochemical elucidation of altered organ and cancer metabolism to develop targeted therapies. When not in lab, Temoc enjoys playing board games and sports with friends and family. He is a recipient of the NIH/NIGMS IMSD and NCI T32 fellowships, and NSF GRFP honorable mention.
Chloe (Eun Jin) Yoo, BA (Postbac Research Student)
Chloe (Eun Jin) is a recent graduate from Vanderbilt University, where she earned her BA in Molecular Cellular Biology. She is planning to attend pharmacy school and is deeply passionate about RNA and its intricate role in cancer biology. In her free time, she enjoys doing pilates, doing Lego, and spending time with her family and friends.
Chloe Heras (Undergraduate Student)
Chloe is a Biological Sciences undergraduate, with a fervid interest in the micro-perspective of cancer mechanisms. She is a recipient of the Rose Hills Foundation and UROP fellowships.
Varvara Rubtsova (Undergraduate Student)
Varvara is an undergraduate student majoring in Biological Sciences. She is interested in nutrient metabolism and public health, and is co-mentored by Dr. Cholsoon Jang; her goal is to understand diet’s influence on health and to translate research discoveries into public health initiatives. In her free time, she enjoys crocheting, climbing, and playing the guitar.
Krystal Huynh (Undergraduate Student)
Krystal is an undergraduate student majoring in Biological Sciences. Her research interest is in delineating the pathological characteristics of cancer progression and regression (supported by UROP fellowship). During her free time, she enjoys baking, playing tennis, and spending time with family and friends.
Alumni
- Lavina Mathur, MS (Life Science Researcher at Stanford University)
- Alexis Anica, BS (PhD Student at Weill Cornell Medicine, Cornell University)
- Lauren Hoffner, BS (PhD Student at Scripps Institute)
Links
- Department of Microbiology & Molecular Genetics
- Cancer Research Institute
- Center for Epigenetics and Metabolism
- Center for Complex Biological Systems
- PhD Program in Cellular & Molecular Biosciences (CMB)
- PhD Program in Mathematical, Computational, and Systems Biology (MCSB)
- MD-PhD Medical Scientist Training Program (MSTP)
Join Us
Our lab is located in Sprague Hall, a modern research building dedicated to cancer investigators. We are seeking highly motivated students and postdocs interested in RNA, signaling, and cancer metabolism to join our research team. Please contact the Principal Investigator at ginalee@uci.edu, with a CV, to inquire about potential opportunities.